3 Interoperability
3.1 Introduction
Ideally, users would be able to leverage the full range of available tools for their analysis. Methods should be selected based on scientific merit (ideally demonstrated by neutral benchmarks), and independent of having been implemented in a given programming language (R or Python) or framework (say, Bioconductor or Seurat).
For single-cell and spatial omics data analysis, being able to leverage different ecosystems is especially powerful. On the one hand, Python offers superb infrastructure for image analysis and machine learning-based approaches. On the other hand, the R programming language has been historically dedicated to statistical computing; as a result, many modern R methods for spatial omics data build on a solid foundation of tools for spatial statistics and statistical modeling in general.
Different data structures, although standardized within a given framework, make switching between languages and tools somewhat cumbersome. In the realm of single-cell and spatial omics, all Bioconductor tools are built around SummarizedExperiment-derived classes, while Seurat, Giotto and VoltRon each rely on own object definitions; Python’s scanpy (Wolf, Angerer, and Theis 2018) and squidpy (Palla et al. 2022) use AnnData. Attempts to alleviate the problem are being made; e.g. zellkonverter and functions from Seurat allow for conversion between Python’s AnnData, R/Bioconductor’s SingleCellExperiment and also SpatialExperiment.
On a higher level, tools that enable interoperability between programming languages have become available. For example, the reticulate package provides an R interface to Python, including support to translate between objects from both languages; basilisk facilitates Python environment management within the Bioconductor ecosystems, but can be interfaced with reticulate as well; and, Quarto can generate dynamic reports from code in different languages.
3.2 Dependencies
3.3 Configuring Python
We first create an environment with Python modules necessary for the examples in this chapter. For that, we first define a basilisk environment and use the resulting conda environment with reticulate.
env <- BasiliskEnvironment(
envname="bkg-interop",
pkgname="base",
channels=c("conda-forge", "bioconda", "pytorch"),
packages=c(
"python=3.11",
"Dask=2024.12.1"),
pip=c(
"squidpy==1.6.2"))
use_virtualenv(obtainEnvironmentPath(env))## Installing pyenv ...
## Done! pyenv has been installed to '/root/.local/share/r-reticulate/pyenv/bin/pyenv'.
## Using Python: /root/.pyenv/versions/3.11.13/bin/python3.11
## Creating virtual environment '/root/.cache/R/basilisk/1.21.5/base/4.5.0/bkg-interop' ...## + /root/.pyenv/versions/3.11.13/bin/python3.11 -m venv /root/.cache/R/basilisk/1.21.5/base/4.5.0/bkg-interop## Done!
## Installing packages: pip, wheel, setuptools## + /root/.cache/R/basilisk/1.21.5/base/4.5.0/bkg-interop/bin/python -m pip install --upgrade pip wheel setuptools## Installing packages: 'Dask==2024.12.1', 'squidpy==1.6.2'## + /root/.cache/R/basilisk/1.21.5/base/4.5.0/bkg-interop/bin/python -m pip install --upgrade --no-user 'Dask==2024.12.1' 'squidpy==1.6.2'## Virtual environment '/root/.cache/R/basilisk/1.21.5/base/4.5.0/bkg-interop' successfully created.3.4 SingleCellExperiment
3.4.1 Calling Python
After configuring the python environment, R commands will now be reticulated; e.g.:
anndata <- reticulate::import("anndata")
example_h5ad <- system.file("extdata", "krumsiek11.h5ad", package = "zellkonverter")
(ad <- anndata$read_h5ad(example_h5ad))## AnnData object with n_obs × n_vars = 640 × 11
## obs: 'cell_type'
## uns: 'highlights', 'iroot'3.4.2 Continuing in R
We can access any of the variables above in R. For basic outputs, this works out of the box:
unique(ad$obs$cell_type)## [1] progenitor Mo Ery Mk Neu
## Levels: Ery Mk Mo Neu progenitorThen again, reticulate supports a few direct type conversions (e.g. dictionary \(\leftrightarrow\) named list). In the example demonstrated here, we can use zellkonverter to go from AnnData to SingleCellExperiment:
(sce <- AnnData2SCE(ad))## class: SingleCellExperiment
## dim: 11 640
## metadata(2): highlights iroot
## assays(1): X
## rownames(11): Gata2 Gata1 ... EgrNab Gfi1
## rowData names(0):
## colnames(640): 0 1 ... 158-3 159-3
## colData names(1): cell_type
## reducedDimNames(0):
## mainExpName: NULL
## altExpNames(0):3.4.3 Back to Python
We can also do the reverse, i.e. go from R’s SingleCellExperiment to Python’s AnnData:
(ad <- SCE2AnnData(sce, X_name="X"))## AnnData object with n_obs × n_vars = 640 × 11
## obs: 'cell_type'
## uns: 'X_name', 'highlights', 'iroot'3.5 SpatialExperiment
Since SpatialExperiment extends the SingleCellExperiment class, conversion operations that we discussed above are also applicable to the SpatialExperiment class. However, to accomplish a full conversion from the AnnData object, we need to manually insert the spatial information using reticulate directly.
3.5.1 Starting with R
For this use case with SpatialExperiment, we will use the dataset from Janesick et al. (2023), which includes Visium measurements on human breast cancer tissue.
id <- "Visium_HumanBreast_Janesick"
pa <- OSTA.data_load(id)
dir.create(td <- tempfile())
unzip(pa, exdir=td)
obj <- TENxVisium(
spacerangerOut=td,
format="h5",
images="lowres")
(spe <- VisiumIO::import(obj))We also have to parse the original scaling information (i.e. scale factor) for spots and images available in standard Visium output. We will use this later during conversion.
We again use the SCE2AnnData function from the previous example, and convert the SingleCellExperiment-relevant components of the SpatialExperiment object to an AnnData object.
(ad <- SCE2AnnData(spe, X_name="counts"))## AnnData object with n_obs × n_vars = 4992 × 18085
## obs: 'in_tissue', 'array_row', 'array_col', 'sample_id'
## var: 'ID', 'Symbol', 'Type'
## uns: 'X_name', 'resources', 'spatialList'
## obsm: 'spatial'We should now populate uns and obsm components of AnnData with spatial coordinates and images. We start with the coordinates.
coords <- spatialCoords(spe)
colnames(coords) <- c("x", "y")
obsm <- list(spatial = coords)Now let’s create the uns component. The list of uns should compose of as many samples as the images in the SpatialExperiment object. Also, each sample entry in the list should have two elements, one for the image and the other for scaling information.
# get image metadata
imgdata <- imgData(spe)
# get image
img <- imgRaster(spe)
img <- apply(img, c(1,2), \(x) col2rgb(x))
img <- aperm(img, perm = c(2,3,1))
img <- img/255
# create uns
uns <- list(
spatial = setNames(list(NULL), imgdata$sample_id)
)
uns[["spatial"]][[imgdata$sample_id]] <-
list(images = list(lowres = img),
scalefactors = scalefactors)Now let’s insert the components to the AnnData object, and write back to an h5ad file.
ad$obs$library_id <- imgdata$sample_id
ad$obsm <- obsm
ad$uns <- uns
ad$write_h5ad("spe.h5ad")3.5.2 Calling Python
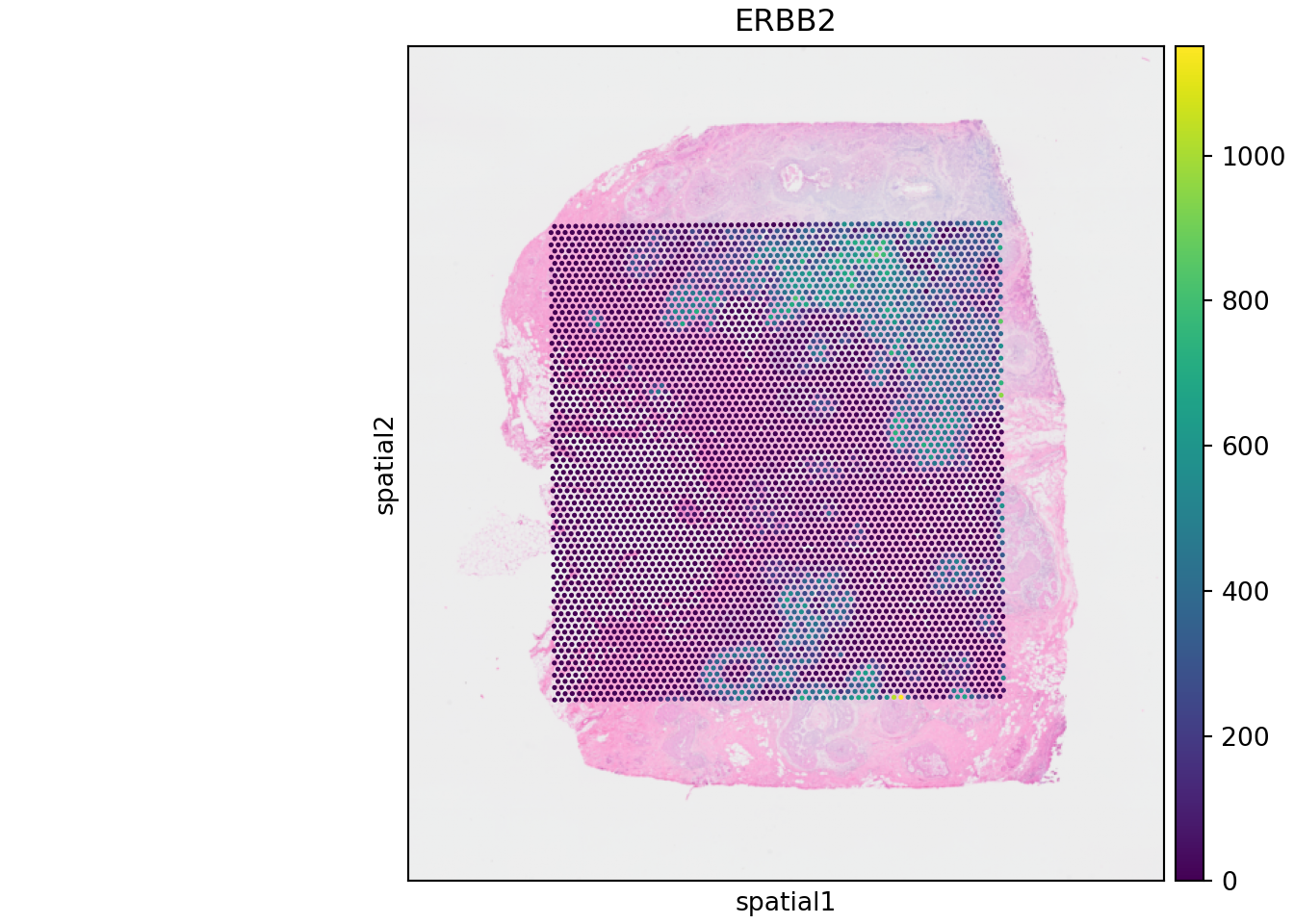
Now that we have converted our SpatialExperiment object to AnnData format, we visualize the Visium data using, for example, the squidpy module.
import squidpy as sq
import anndata as ad
import matplotlib.pyplot as pltNow, we read spe.h5ad and visualize features and/or gene expression (e.g. ERBB2).
adata = ad.read_h5ad("spe.h5ad")
adata.var_names = adata.var['Symbol'].astype(str)
sq.pl.spatial_scatter(adata,
color=["ERBB2"],
img_res_key = "lowres")