SpatialExperiment data class for storing and manipulating spatial transcriptomics datasets within the Bioconductor framework.Bioconductor provides several data classes for storing and manipulating spatial transcriptomics datasets. By relying on these consistent data structures, we can easily connect methods and packages developed by different research groups to build analysis workflows that include the latest state-of-the-art methods.
Below, we describe the Bioconductor data classes used in this book.
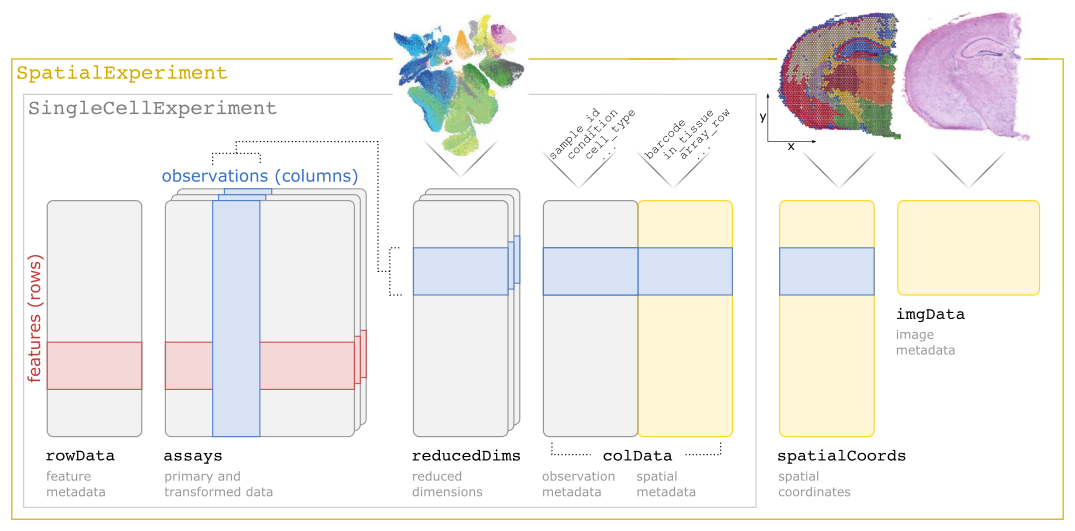
The SpatialExperiment class (Righelli et al. 2022) is the core data class used in this book. This class allows us to store datasets at the spot or cell level, i.e. data from sequencing-based platforms at the spot level, or data from imaging-based platforms aggregated to the cell level.
SpatialExperiment builds on the SingleCellExperiment class (Amezquita et al. 2020) for single-cell RNA sequencing data, with additional customizations to store spatial information, such as spatial coordinates and image files.
A summary of the SpatialExperiment object structure is shown in the following schematic. Briefly, a SpatialExperiment object consists of (i) assays containing expression counts, (ii) rowData containing information on features, i.e. genes, (iii) colData containing information on spots or cells, including nonspatial and spatial metadata, (iv) spatialCoords containing spatial coordinates, and (v) imgData containing image data. For spot-based data, a single assay named counts is used.
For more details, see the SpatialExperiment package Bioconductor vignette or the paper describing the class (Righelli et al. 2022).

SpatialExperiment data class for storing and manipulating spatial transcriptomics datasets within the Bioconductor framework.Imaging-based spatial transcriptomics datasets may contain additional information, including the spatial coordinates of individual mRNA molecules, and segmentation boundaries for cells or nuclei.
The following Bioconductor classes provide additional functionality to store and manipulate this information.
These classes extend SpatialExperiment and are compatible with SpatialExperiment for analyses at the aggregated cell level.
SpatialFeatureExperiment is available as a Bioconductor package and described by the authors in their paper (Moses et al. 2023).
MoleculeExperiment is available as a Bioconductor package and described by the authors in their paper (Peters Couto et al. 2023).